Note
Go to the end to download the full example code
Tutorial for basic usage¶
[[-3.14159265 -1. 2. ]
[-3.14159265 -0.5 2. ]
[-3.14159265 0. 2. ]
[-3.14159265 0.5 2. ]
[-3.14159265 1. 2. ]
[-2.74889357 -1. 1.27648914]
[-2.74889357 -0.5 1.707008 ]
[-2.74889357 0. 1.94618514]
[-2.74889357 0.5 1.99402057]
[-2.74889357 1. 1.85051428]
[-2.35619449 -1. 0.66312624]
[-2.35619449 -0.5 1.45862137]
[-2.35619449 0. 1.90056311]
[-2.35619449 0.5 1.98895146]
[-2.35619449 1. 1.72378641]
[-1.57079633 -1. 0.109375 ]
[-1.57079633 -0.5 1.234375 ]
[-1.57079633 0. 1.859375 ]
[-1.57079633 0.5 1.984375 ]
[-1.57079633 1. 1.609375 ]
[-0.78539816 -1. 0.66312624]
[-0.78539816 -0.5 1.45862137]
[-0.78539816 0. 1.90056311]
[-0.78539816 0.5 1.98895146]
[-0.78539816 1. 1.72378641]
[-0.39269908 -1. 1.27648914]
[-0.39269908 -0.5 1.707008 ]
[-0.39269908 0. 1.94618514]
[-0.39269908 0.5 1.99402057]
[-0.39269908 1. 1.85051428]
[ 0. -1. 2. ]
[ 0. -0.5 2. ]
[ 0. 0. 2. ]
[ 0. 0.5 2. ]
[ 0. 1. 2. ]
[ 0.39269908 -1. 2.72351086]
[ 0.39269908 -0.5 2.292992 ]
[ 0.39269908 0. 2.05381486]
[ 0.39269908 0.5 2.00597943]
[ 0.39269908 1. 2.14948572]
[ 0.78539816 -1. 3.33687376]
[ 0.78539816 -0.5 2.54137863]
[ 0.78539816 0. 2.09943689]
[ 0.78539816 0.5 2.01104854]
[ 0.78539816 1. 2.27621359]
[ 1.57079633 -1. 3.890625 ]
[ 1.57079633 -0.5 2.765625 ]
[ 1.57079633 0. 2.140625 ]
[ 1.57079633 0.5 2.015625 ]
[ 1.57079633 1. 2.390625 ]
[ 2.35619449 -1. 3.33687376]
[ 2.35619449 -0.5 2.54137863]
[ 2.35619449 0. 2.09943689]
[ 2.35619449 0.5 2.01104854]
[ 2.35619449 1. 2.27621359]
[ 2.74889357 -1. 2.72351086]
[ 2.74889357 -0.5 2.292992 ]
[ 2.74889357 0. 2.05381486]
[ 2.74889357 0.5 2.00597943]
[ 2.74889357 1. 2.14948572]
[ 3.14159265 -1. 2. ]
[ 3.14159265 -0.5 2. ]
[ 3.14159265 0. 2. ]
[ 3.14159265 0.5 2. ]
[ 3.14159265 1. 2. ]]
Coefficients all same? True
Knots all same? True
Antiderivative of derivative:
Coefficients differ by constant? True
Knots all same? True
Derivative of antiderivative:
Coefficients the same? True
Knots all same? True
8 import ndsplines
9 import numpy as np
10 import matplotlib.pyplot as plt
11
12 # generate grid of independent variables
13 x = np.array([-1, -7/8, -3/4, -1/2, -1/4, -1/8, 0, 1/8, 1/4, 1/2, 3/4, 7/8, 1])*np.pi
14 y = np.array([-1, -1/2, 0, 1/2, 1])
15 meshx, meshy = np.meshgrid(x, y, indexing='ij')
16 gridxy = np.stack((meshx, meshy), axis=-1)
17
18
19 # generate denser grid of independent variables to interpolate
20 sparse_dense = 2**7
21 xx = np.concatenate([np.linspace(x[i], x[i+1], sparse_dense) for i in range(x.size-1)]) # np.linspace(x[0], x[-1], x.size*sparse_dense)
22 yy = np.concatenate([np.linspace(y[i], y[i+1], sparse_dense) for i in range(y.size-1)]) # np.linspace(y[0], y[-1], y.size*sparse_dense)
23 gridxxyy = np.stack(np.meshgrid(xx, yy, indexing='ij'), axis=-1)
24
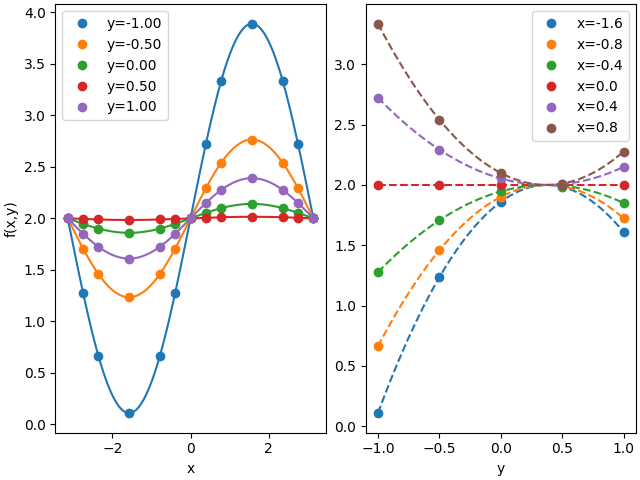
25 def plots(sparse_data, dense_data, ylabel='f(x,y)'):
26 fig, axes = plt.subplots(1, 2, constrained_layout=True)
27 for yidx in range(sparse_data.shape[1]):
28 axes[0].plot(x, sparse_data[:, yidx], 'o', color='C%d'%yidx, label='y=%.2f'%y[yidx])
29 axes[0].plot(xx, dense_data[:, np.clip(yidx*sparse_dense, 0, yy.size-1)], color='C%d'%yidx)# label='y=%.1f'%y[yidx])
30
31 axes[0].legend()
32 axes[0].set_xlabel('x')
33 axes[0].set_ylabel(ylabel)
34 for xidx in range(sparse_data.shape[0]//2):
35 axes[1].plot(yy, dense_data[(xidx+3)*sparse_dense, :], '--', color='C%d'%xidx,)# label='x=%.1f'%x[xidx+3])
36 axes[1].plot(y, sparse_data[xidx+3, :], 'o', color='C%d'%xidx, label='x=%.1f'%x[xidx+3],)
37
38 axes[1].legend()
39 axes[1].set_xlabel('y')
40 plt.show()
41
42 # evaluate a function to interpolate over input grid
43 meshf = np.sin(meshx) * (meshy-3/8)**2 + 2
44
45 # create the interpolating splane
46 interp = ndsplines.make_interp_spline(gridxy, meshf)
47
48 # evaluate spline over denser grid
49 meshff = interp(gridxxyy)
50
51
52 plots(meshf, meshff)
53
54
55 ##
56
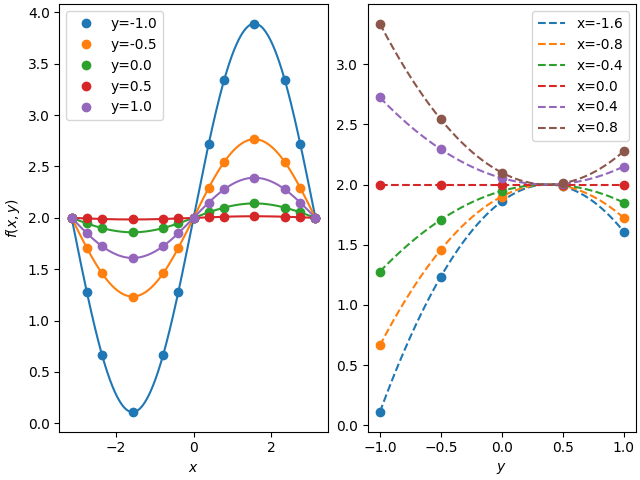
57 # as subplots
58 fig, axes = plt.subplots(1,2, constrained_layout=True)
59
60 gridxxy = np.stack(np.meshgrid(xx, y, indexing='ij'), axis=-1)
61 meshff = interp(gridxxy)
62
63 for yidx in range(meshf.shape[1]):
64 axes[0].plot(x, meshf[:, yidx], 'o', color='C%d'%yidx, label='y=%.1f'%y[yidx])
65 axes[0].plot(xx, meshff[:, yidx], color='C%d'%yidx)
66 axes[0].legend()
67 axes[0].set_xlabel('$x$')
68 axes[0].set_ylabel('$f(x,y)$')
69
70 # y-dir plot
71 gridxyy = np.stack(np.meshgrid(x, yy, indexing='ij'), axis=-1)
72
73 meshff = interp(gridxyy)
74 for xidx in range(meshf.shape[0]//2):
75 axes[1].plot(yy, meshff[xidx*1+3, :], '--', color='C%d'%xidx, label='x=%.1f'%x[xidx*1+3])
76 axes[1].plot(y, meshf[xidx*1+3, :], 'o', color='C%d'%xidx)
77
78 axes[1].legend()
79 axes[1].set_xlabel('$y$')
80 # plt.ylabel(r'$\frac{\partial f(x,y)}{\partial y}$')
81 plt.show()
82
83 ##
84
85 # we could also use tidy data format to make the grid
86
87 tidy_data = np.dstack((gridxy, meshf)).reshape((-1,3))
88 print(tidy_data)
89
90 tidy_interp = ndsplines.make_interp_spline_from_tidy(tidy_data, [0,1], [2])
91
92 print("\nCoefficients all same?", np.all(tidy_interp.coefficients == interp.coefficients))
93 print("Knots all same?", np.all([np.all(knot0 == knot1) for knot0, knot1 in zip(tidy_interp.knots, interp.knots)]))
94
95 # send to example of least squares
96 ##
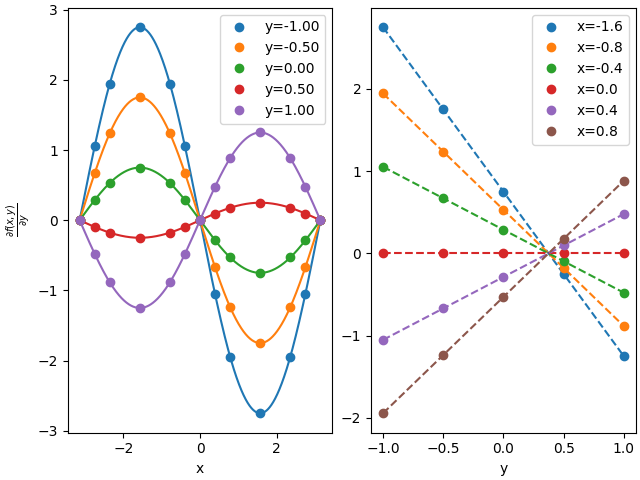
97 # two ways to evaluate derivative - y direction
98
99 deriv_interp = interp.derivative(1)
100 deriv1 = deriv_interp(gridxy)
101 deriv2 = interp(gridxxyy, nus=np.array([0,1]))
102
103 plots(deriv1, deriv2, r'$\frac{\partial f(x,y)}{\partial y}$')
104
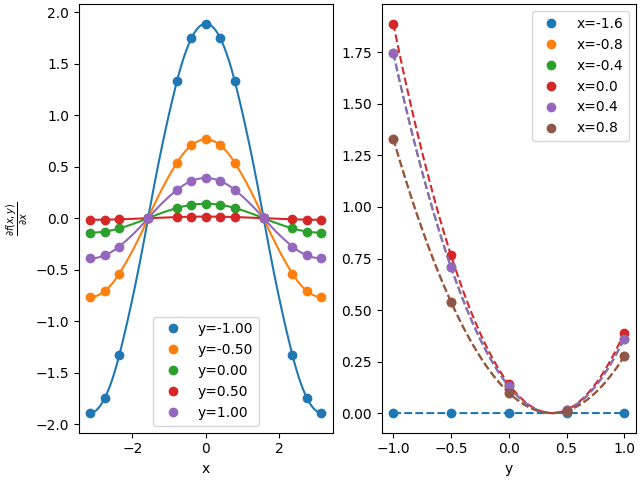
105 ##
106 # two ways to evaluate derivatives x-direction: create a derivative spline or call with nus:
107 deriv_interp = interp.derivative(0)
108 deriv1 = deriv_interp(gridxy)
109 deriv2 = interp(gridxxyy, nus=np.array([1,0]))
110
111 plots(deriv1, deriv2, r'$\frac{\partial f(x,y)}{\partial x}$')
112 ##
113
114 # Calculus demonstration
115 interp1 = deriv_interp.antiderivative(0)
116 coeff_diff = interp1.coefficients - interp.coefficients
117 print("\nAntiderivative of derivative:\n","Coefficients differ by constant?", np.allclose(interp1.coefficients+2.0, interp.coefficients))
118 print("Knots all same?", np.all([np.all(knot0 == knot1) for knot0, knot1 in zip(interp1.knots, interp.knots)]))
119
120 antideriv_interp = interp.antiderivative(0)
121
122 interp2 = antideriv_interp.derivative(0)
123 print("\nDerivative of antiderivative:\n","Coefficients the same?", np.allclose(interp2.coefficients, interp.coefficients))
124 print("Knots all same?", np.all([np.all(knot0 == knot1) for knot0, knot1 in zip(interp2.knots, interp.knots)]))
Total running time of the script: (0 minutes 2.784 seconds)